Principal Component Analysis (PCA)
Sumber: http://mengnote.blogspot.com/2013/05/an-intuitive-explanation-of-pca.html
Many research papers apply PCA (Principal Component Analysis) to their data and present results to readers without further explanation of the method. When people search on the internet for a definition of PCA, they sometimes get confused, often by terms like "covariance matrix", "eigenvectors" or "eigenvalues". It is not surprising because most explanatory articles focus on detailed calculation process instead of the basic idea of PCA. They are mathematically correct, yet often not intuitively readable at first glance.
For a mathematical method, I believe most people only need to understand the logic and limitations of it and let software packages to do the rest (implementation, calculation, etc.). Here I am trying to explain that PCA is not an impenetrable soup of acronyms but a quite intuitive approach. It can make large-scale data "smaller" and easier to handle.
PCA is nothing but coordinate system transformation: A simple example
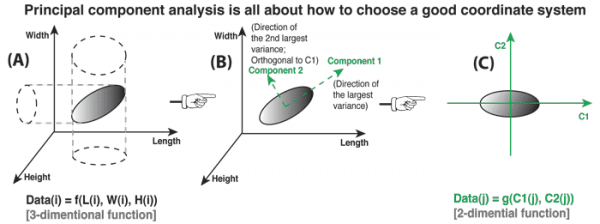
Figure 1. (A) A simple object described by a complex set of variables (or coordinate system). (B) Find new variables (coordinate axes) orthogonal to each other and pointing to the direction of largest variances. (C) Use new variables (coordinate axes) to describe object in a more concise way.
A simple object could be described by complex and unnecessary variables. We don't intend to do so, but often can not avoid that. One problem is how to find the simplest way to describe an object? Here is an example to show that the answer could be by choosing a proper coordinate system.
Let's assume that we have a "3-D shape detect machine" that can scan anything in front it and output a model of that object. We use it to scan a ellipse made of paper (paper thickness can be neglected). The output model uses three axes: L (Length), W (Width) and H (Height) that perpendicular to each other to represent the 3-D world (see Figure 1A). So each data point on that object can be written as a function of three variables:
Data(i) = f(L(i), W(i), H(i)) [function 1]
Apparently, this is not the best way to describe an ellipse. I guess most people will do the following improvements:
Find the geometric center of the ellipse, and set it as the coordinate origin; Find the direction along which the ellipse has the longest radius. We call this direction (or variable along this direction) "component one". Find another direction that perpendicular to the first one, and along which the ellipse has the second longest radius. We call this direction "component two". (step 1~3, see Figure 1B) Re-plot the ellipse under the new coordinate system defined by component one (C1) and two (C2). (see Figure 1C)
In the new coordinate system, each data point on that ellipse can be re-written as a function of two variables:
Data(j) = g(C1(j), C2(j)) [function 2]
Now it is quite clear that, after proper coordinate system transform, we get:
Fewer variables (or lower dimensions of variables) of function 2 compared to function 1.
(L, W, H) --> (C1, C2)
No information lost.
function 1 == function 2
The relative geometric positions of all data points remain unchanged.
That's exactly what PCA analysis do. The term "principal component" denotes new variables (or coordinate systems) we choose to describe our data in a more concise or convenient way. All principal components must satisfy two conditions:
They must be perpendicular (or mathematically, Orthogonal) to each other.
This means principal components are NOT linear correlated between each other.
And that's why PCA can reduce the number of variables with out losing information: variables in raw data are not independent, and correlated variables cause redundancy.
Numerical IDs of components are ordered by the length of radius (mathematically speaking, variance) of our data points along them. So our data must have the largest variance along the axes of component 1, and the 2nd largest variance along the axes of component 2, and so on.
This means the higher order a component have, the more important it is.
Some times we may sacrifice minor components to further reduce the number of variables. eg, If the first two principal components (out of the total 10) contributed 90% of variance of the data, we might like to focus on them only and discard the other 8 components.
Why we consider the axes along which the data has the biggest variance as the most important one? The reason is that along that axes we can get the best separation of data points. A more natural explanation could be like: along that axes we can see the largest divergence from the mean value. Just like when we talk about trees, rail ways and stars, we describe them as tall, long and far -- the dimensions that have greatest divergences.
PCA can reduce number of variables by eliminating correlations between them: A more realistic example Figure 2 (A) Data set of gene expression level in different tissues. (B) Gene expression level is a n-dimensional function. (C) PCA process. (D) After PCA, gene expression level becomes a k-dimensional function (k<n). (E) Data set is "compressed" with no information lost. Suppose we measure expression levels of N genes in M tissues. We will get a data set of M*N matrix (see Figure 2A). Mathematically, we can describe the data set as:
Tissue(i) = f(gene1(i), gene2(i)...geneN(i))
In which Tissue(i) means expression level of all genes in the i-th tissue, and gene1(i) denotes expression level of gene 1 in the i-th tissue, and so on. As this function has N variables, we can not use a graph to show the N-dimensional function when N>3. But we can imagine the high dimension as multiple coordinate axes (see Figure 2B). Each axes stands for one gene's expression level.
We use PCA to reduce the dimension of variables:
Find the mean value of all data, and set it as the coordinate origin; Find the direction along which the data set has the largest variance. We call it "component 1". Find another direction that orthogonal to all components already found, and along which the data has the longest possible variance. We call this direction "component 2". repeat step 3 until we get K principal components and no more component can be found. (step 1~4, see Figure 2C) Re-plot the data set under the new coordinate system defined by K principal components (see Figure 2D).
Note: These steps are conceptual framework used only for understanding PCA. In reality we use linear algebra operations of the same idea but may have quite different appearance. Now we reduced the dimension of variables from N to K and K<N (see Figure 2E). A example provided by Nature Biotechnology 26, 303 - 304 (2008) that when N = 8,534 , K = 104. This means a huge amount of redundancy was removed from original expression data set. Imagine the difference between two functions that have 8,534 and 104 variables accordingly. PCA can make large scale data much easier to handle. Original data table (Figure 2A) will be greatly compressed (Figure 2E) after PCA process.
PCA can reduce data dimension at a minimum cost. But it can not do anything other than that. we still need to use specific approaches for next step analyses like clustering, inferring or other jobs.
PCA has limitations : example of failures Figure 3. (A) PCA on non-Gaussian source data. (B) Principal components given by PCA may still be correlated in a non-linear way. Prior knowledge could help us to find out the best principal components. Any algorithm could fail when its assumptions are not satisfied. PCA makes "the largest variance" assumption. If the data does not follow a multidimensional normal (Gaussian) distribution, PCA may not give the best principal components (see Figure 3A). For non-Gaussian source data, we will need independent component analysis (ICA) to separate data into additive, mutual statistical independent sub-components.
PCA also makes assumption that all principal components are orthogonal (linear uncorrelated) to each other. So it may fail when principal components are correlated in a non-linear manner. For a ring-shape data set (see Figure 3B), PCA will give two principal components C1 and C2, which are actually correlated through rotation angle θ. It's easy to know that instead of C1 or C2, θ should be the real first-order principal component. In such cases, before applying PCA, we need to first perform a non-linear transformation on raw data to create a new linear space in which variable θ becomes a linear dimension. And this method was called kernel-PCA.
PCA implementations : eigenvalue decomposition (EVD) and singular value decomposition (SVD) Figure 4 As a conceptual framework, PCA could have different implementations. May be the most popular ones are eigenvalue decomposition (EVD) and singular value decomposition (SVD). They both can be used to find principal components and will give same results in most cases.
Before applying EVD or SVD, we should define our goal as to find a proper transformation through which a NxM matrix can be transformed (or compressed) into a NxK (K<=M) matrix with no information lost (see Figure 4A).
Now let's imagine EVD algorithm as a machine with inputs and outputs. We feed EVD with our raw data set X, a NxM matrix, as input. EVD will output the following (see Figure 4B):
Two new matrices W and D. W contains all principal component vectors, while D contains all ranks of those vectors (ordered from the largest variance to the least one). XT
and WT are transposes of X and W accordingly. So they are not considered as new. An equation : XXT=WDWT
We know that WD is a NxK (K<=M) matrix which is exactly of the same format we want to transform our data set into. So we can re-write the equation into:
XXTW=WD[Solution1]
OK, we get a solution (transformation) for our goal using EVD. Because we can transform a NxM matrix X into a NxK (K<=M) matrix WD (see Figure 4C).
SVD is another method that can be used to reach our goal. Let's imagine SVD as a machine with inputs and outputs. We feed SVD with data set X, the NxM matrix, as input. SVD will output the following (see Figure 4D):
Three new matrix U, Σ
and VT. U and VT contain principal component vectors for two directions (column and row of raw data) accordingly. Σ contains ordered ranks of those principal components. An equation : X=UΣVT
We know that UΣ is a NxK (K<=M) matrix which is exactly of the same format we want to transform our data set into. We can re-write the equation into:
XV=UΣ[Solution2]
So we get another solution for our goal using SVD. Because we can transform a NxM matrix X into a NxK (K<=M) matrix UΣ (see Figure 4E).
Another question is, as EVD and SVD seemingly do the same work, which one should we choose? I personally recommend SVD. The reason is EVD requires calculation of XXT which might lead to losing of precision when X is a Läuchli matrix. In this scenario, XXT will perform operations of adding very large numbers with very small numbers, and the small ones will be "eaten" by large ones. SVD does not have this problem.