Orange: DBSCAN
Sumber: https://docs.biolab.si//3/visual-programming/widgets/unsupervised/DBSCAN.html
Groups items using the DBSCAN clustering algorithm.
Input
Data: input dataset
Output
Data: dataset with cluster index as a class attribute
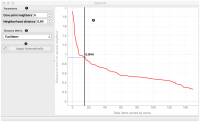
The widget applies the DBSCAN clustering algorithm to the data and outputs a new dataset with cluster indices as a meta attribute. The widget also shows the sorted graph with distances to k-th nearest neighbors. With k values set to Core point neighbors as suggested in the methods article. This gives the user the idea of an ideal selection for Neighborhood distance setting. As suggested by authors this parameter should be set to the first value in the first “valley” in the graph.
- Set minimal number of core neighbors for a cluster and *maximal neighborhood distance.
- Set the distance metric that is used in grouping the items.
- If Apply Automatically is ticked, the widget will commit changes automatically. Alternatively, click Apply.
- The graph shows the distance to the k-th nearest neighbor. k is set by the Core point neighbor option. With moving the black slider left and right you can select the right Neighbourhood distance.
Contoh
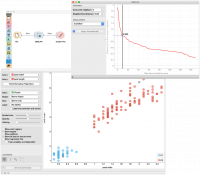
In the following example, we connected the File widget with selected Iris dataset to the DBSCAN widget. In the DBSCAN widget, we set Core points neighbors parameter to 5. And select the Neighbourhood distance to the value in the first “valley” in the graph. We show clusters in the Scatter Plot widget.